Trying the Program Online
You can explore the program online by clicking here. This link takes you to the graphical user interface of the program. There, you can try various available examples for building DNA, RNA, and other alternative nucleic acids. You can also explore how the structure and energy of nucleic acids change as you play with the helical parameters. Additionally, you can upload a three-dimensional structure of an alternative nucleic acid backbone or nucleobase construct a new nucleic acid analogs. See the Examples section to learn how to use the grahpical interface.
This online version is useful for testing the program. The online version uses free cloud computing resources provided by Binder, and this comes with several limitations. If your session remains inactive for 10-20 minutes, your session will be terminated. Moreover, the cloud computers limit how long you can remain in a session. To prevent the loss of data when the session terminates because of inactivity, the program automatically download the output files after it finishes running. You may need to adjust the settings of the browser to allow pop-up windows to open for downloading.
If you need a more permanent version of the program, you can install it in your computer.
Installation
The pNAB program is available as a conda package. Download the light-weight miniconda or the more fully featured Anaconda. Open the conda terminal and type:
This creates and activates a new conda environment called pnab.
- Note
- You may need to activate the NGLView library to visualize molecules: jupyter-nbextension enable nglview --py --sys-prefix
-
In Windows, the program may not compute energires correctly if the environment variable BABEL_DATADIR is not set. Set the variable to thefolder inside your Miniconda/Anaconda folder.share\openbabel
Running the Program
The program can be used through a graphical user interface or a simple python script.
Using the Graphical User Interface
From the conda command-line, run:
Alternatively, open Anaconda Navigator (details):
In the Jupyter Notebook tab, click on Launch. If a browser does not open automatically, you may need to open it this way:
When a browser opens with the Jupyter page, it displays the folders in your computer. navigate to an appropriate folder or create a new folder if needed, then click on New > python 3
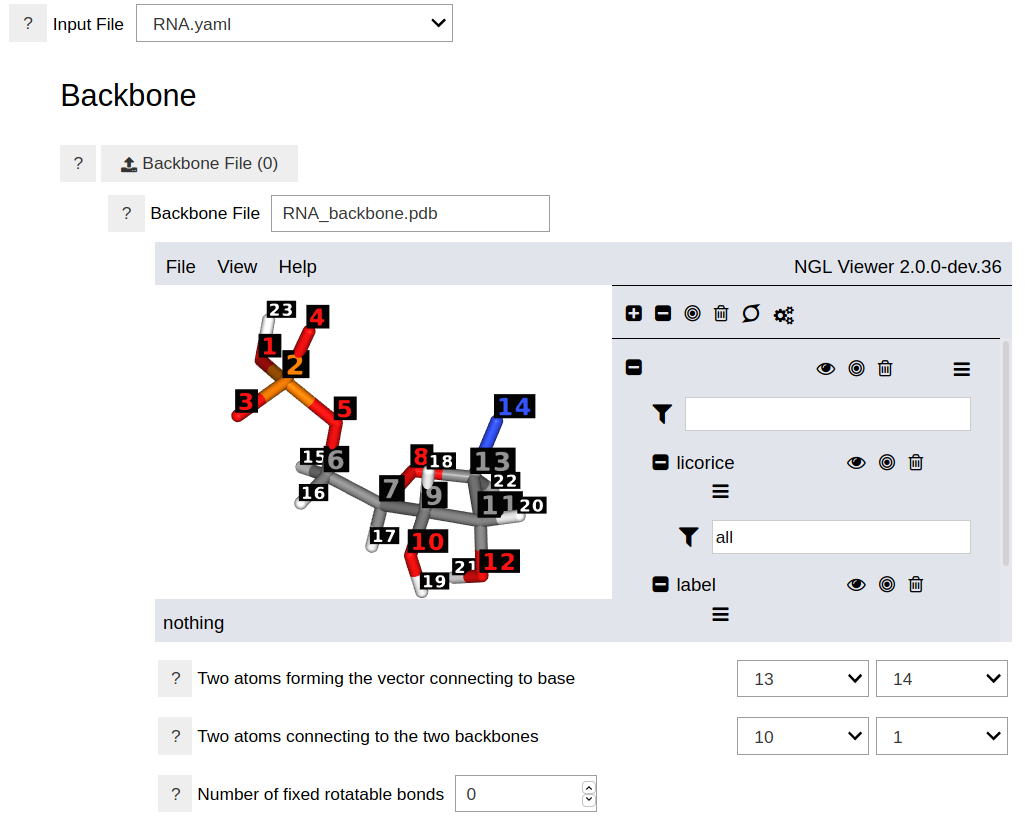
This opens an empty notebook. Type the the following two lines in the first cell and run it. This should show the graphical user interface of the program.
Using a Python Script
The following python script would run the program to generate an RNA structure. The file RNA.yaml is an example file available in the program.
To create your own input files, you can use the installed grahpical user interface or the online version. Alternatively, you can write an input file using a text editor and run it. See the Examples section to learn the content of the input files.
 1.8.18
1.8.18